Integrative Genomics Viewer (IGV) in OOD
The IGV is a high-performance, easy-to-use, interactive tool for the visual exploration of genomic data.
Please see the following links for details:
Getting Started
You can run IGV in OOD by going to the URL ood.torch.hpc.nyu.edu in your browser and selecting IGV from the Interactive Apps pull-down menu at the top of the page. Once you've used it and other interactive apps they'll show up on your home screen under the Recently Used Apps header.
Be aware that when you start from Recently Used Apps it will start with the same configuration that you used previously. If you'd like to configure your IGV session differently, you'll need to select it from the menu.
Configuration
You can select the number or cores, amount of memory, amount of time, and optional Slurm options.
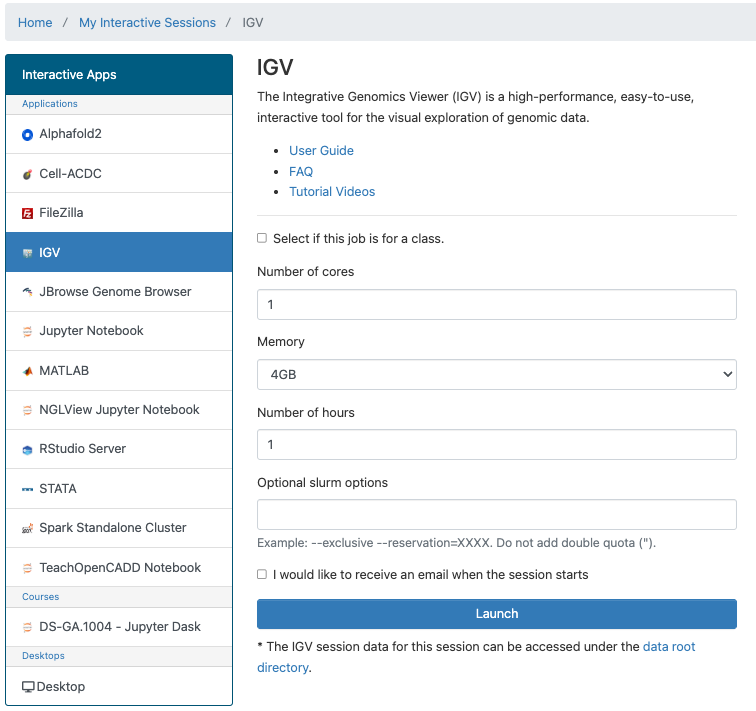
IGV running in OOD
After you hit the Launch button you'll have to wait for the scheduler to find you node(s) to run on:
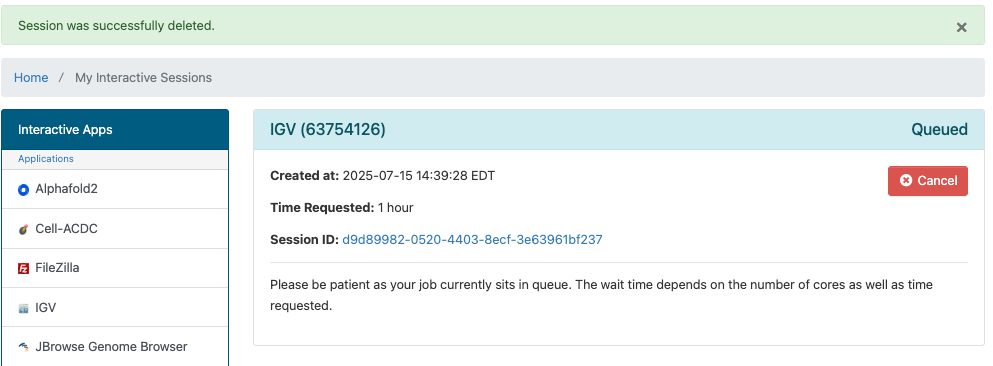
Then you'll have a short wait for IGV itself to start up.
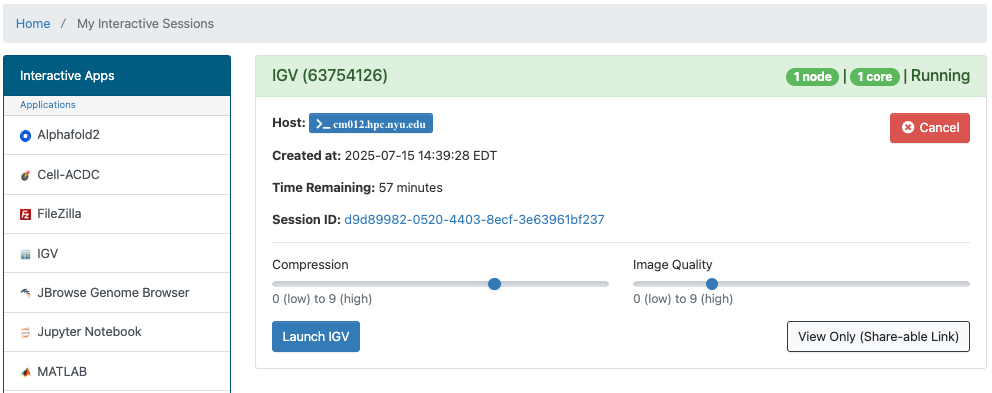
Once that happens you'll get one last form that will allow you to:
- make changes to compression and image qualtiy
- open a terminal window on the compute node your IGV session is running on
- get a link that you can share that will allow others to view your IGV session
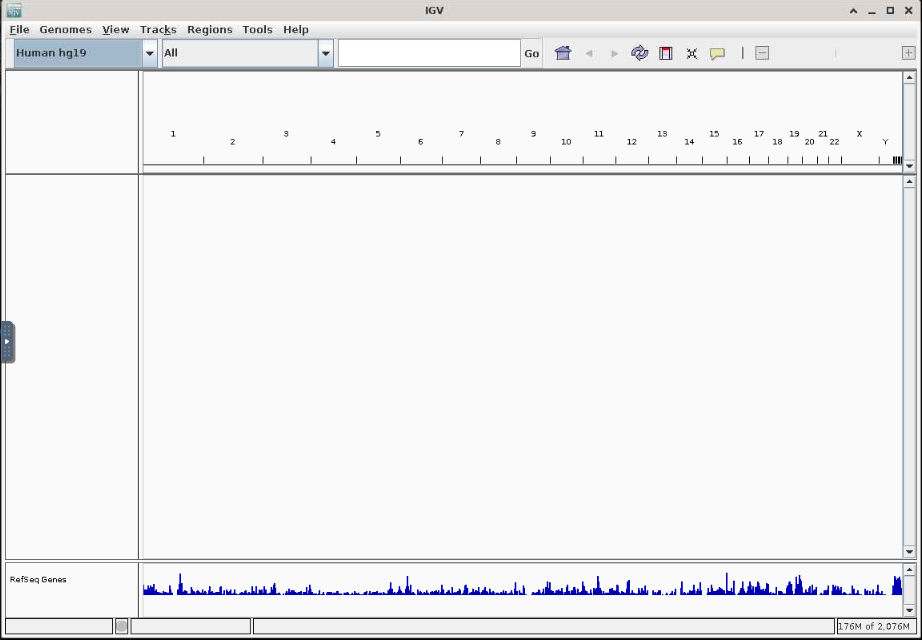
Then after you hit the Launch IGV button you'll have the familiar IGV interface to use.